Bioschemas, what and why?
In this tutorial you will learn what Bioschemas is, what the added value to schema.org.is and what the main elements in Bioschemas are
1. What is Bioschemas?
Bioschemas is a community project built on top of schema.org, aiming to improve interoperability in Life Sciences so resources can better communicate and work together by using a common markup on their websites.
Using schema.org markup on web pages enables the generation of ‘info box’ summaries in typical web search results pages, as exemplified by google search results. Bioschemas aims to make it possible to get similar summaries, but focused on Life Science resources such as Proteins, Samples, Beacons, Tools, Training, Life science events and so on.
Imagine an insulin summary appearing within search results, but rather than pointing to Wikipedia, that summary would direct one to specialized resources such as Orphanet or CATH as seen in Figure 1. In this way you would get a quick overview while also being provided links to relevant resources, all in one search.
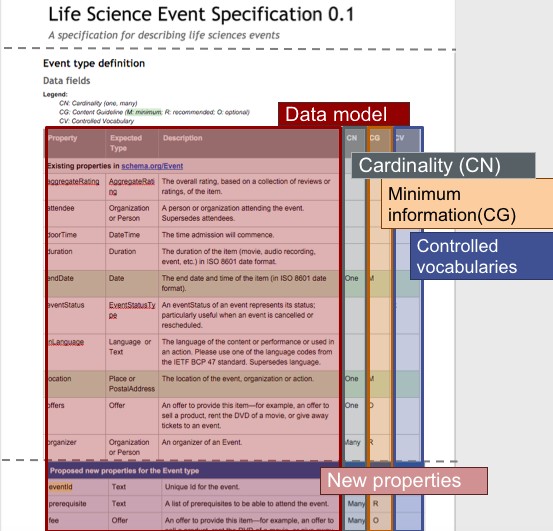
2. What are the benefits of Bioschemas?
Bioschemas inherits the benefits from schema.org, i.e., enabling machines to understand what your metadata is in advance, making it easier to find, integrate, and re-use your data. It also brings some benefits tailored to the Life Sciences community. In Figure 2, you can find a graphical summary of such benefits, which are explained in more detail in the paragraphs below.


Disclaimer: Initially, Bioschemas types were developed with the aim to eventually mature those types and have them integrated for direct use in schema.org. While this remains desirable, it is not essential; community tools and resources are being developed to directly harvest this markup, and there are activities in progress to migrate Bioschemas markup from individual resources to the EOSC (European Open Science Cloud).
- Bioschemas focuses on key properties prioritised as Minimum, Recommended and Optional based on community agreements and common practices

- Minimum properties should be provided
- Recommended properties should be provided whenever possible and available
- Optional properties could be omitted unless important or relevant for your resource
e.g., For the Event case shown on Figure 2, endDate and location are minimum while organizer is recommended.
Reminder: a property helps you describe your resource
- Bioschemas provides additional recommendations regarding properties cardinality

e.g., For the Event case, endDate should be ONE while organizer could be MANY
- Bioschemas customises schema.org types (see previous tutorial) to better supports needs on the life sciences community

- Bioschemas reuses terms from well-known ontologies, thus avoiding reinventing the wheel

Protein, a BioChemEntity profile, includes some properties that come from well-known ontologies. For instance associatedWith comes from from the Sequence Ontology. By reusing terms, Bioschemas aims to avoid reinventing the wheel.
Keywords: schemaorg, markup, structured data, bioschemas
Topics:
Audience:
- (General interest) People interested in introductory information to Bioschemas
Authors:
Contributors:
License: CC-BY 4.0
Version: 2.0
Last Modified: 17 February 2021

