How to check your Bioschemas deployment
This guide will show you how to check your deployed markup, both locally and what is retrieved by external services.
1. Using a browser to check your deployment
There are a few different ways to check that markup is being included on your webpage; regardless of whether the markup is rendered server side or injected client-side through Javascript. One of the simplest is to use your browser to inspect the rendered page content.
In Firefox right click on the page and select ‘Inspect Element’, in Chrome it is the ‘Inspect’ option. You can then search within the generated HTML to look for ‘schema.org’. This is depicted in the screenshot below for the DisProt protein DP00003.
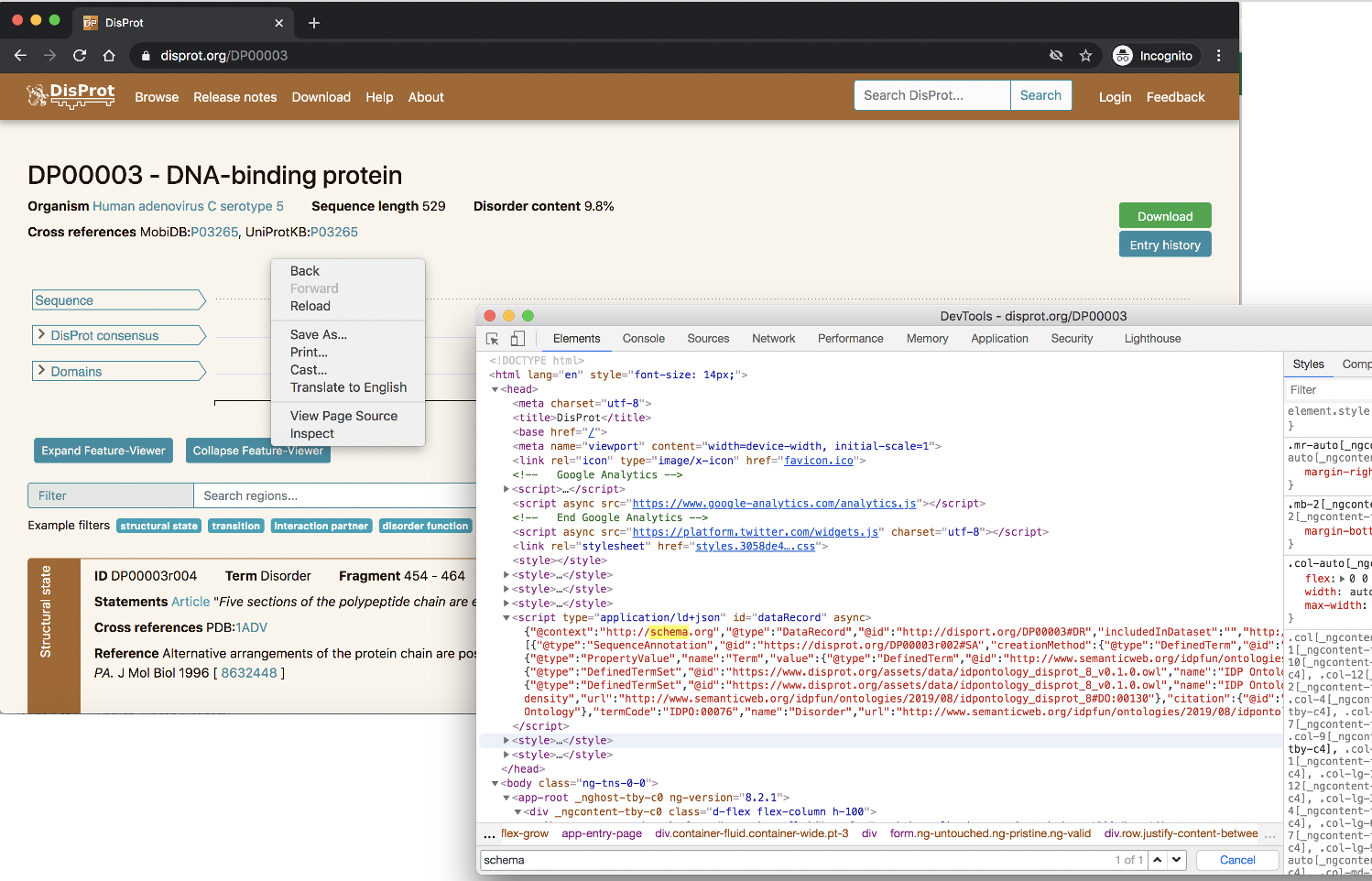
2. Discovering what markup external services can retrieve
Once you have verified that the content is rendered on the page, you can test what external services are able to pick up. Some of these services simply retrieve the markup while others will validate it against a defined profile.
Note that since Bioschemas is an evolving extension of Schema.org, not all types and properties will be known to all services. Additionally, only Bioschemas specific services will know about the Bioschemas profiles; proprietary services will validate against their own internal views of Schema.org.
2.1. Google Structured Data Testing Tool
https://search.google.com/structured-data/testing-tool
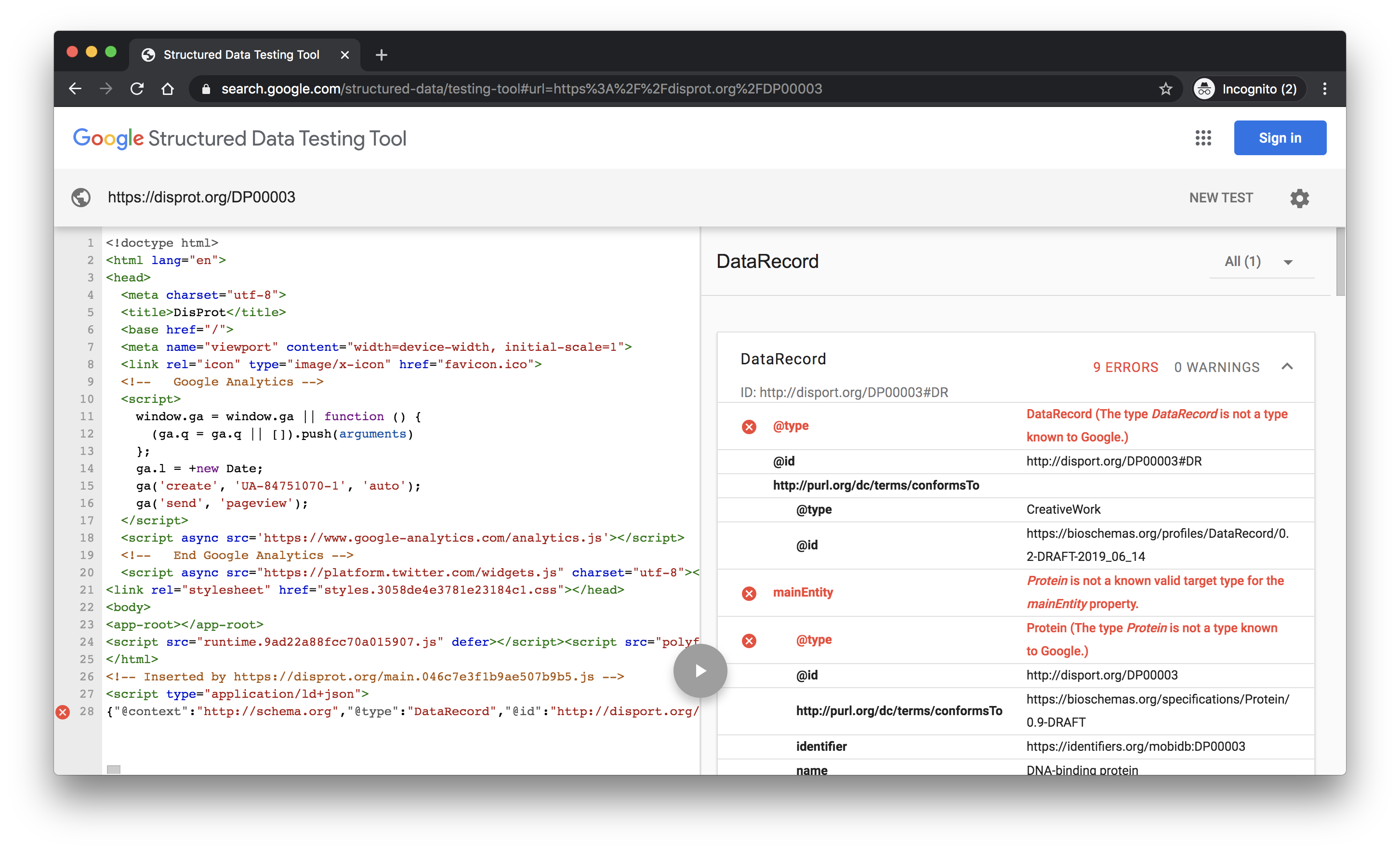
This site gives a nice hierarchical rendering of the discovered content and it can detect markup from Single Page Application engines. The site knows about the core Schema.org vocabulary and the properties that Google use within certain types for their search results. It does not know about the Bioschemas proposals for Schema.org types and properties or our profiles. As a result, you will get warnings and errors for using Bioschemas types.
The screenshot shows the tool results for the DisProt protein DP00003. Click here to see the results.
2.2. Google Rich Results Test
https://search.google.com/test/rich-results
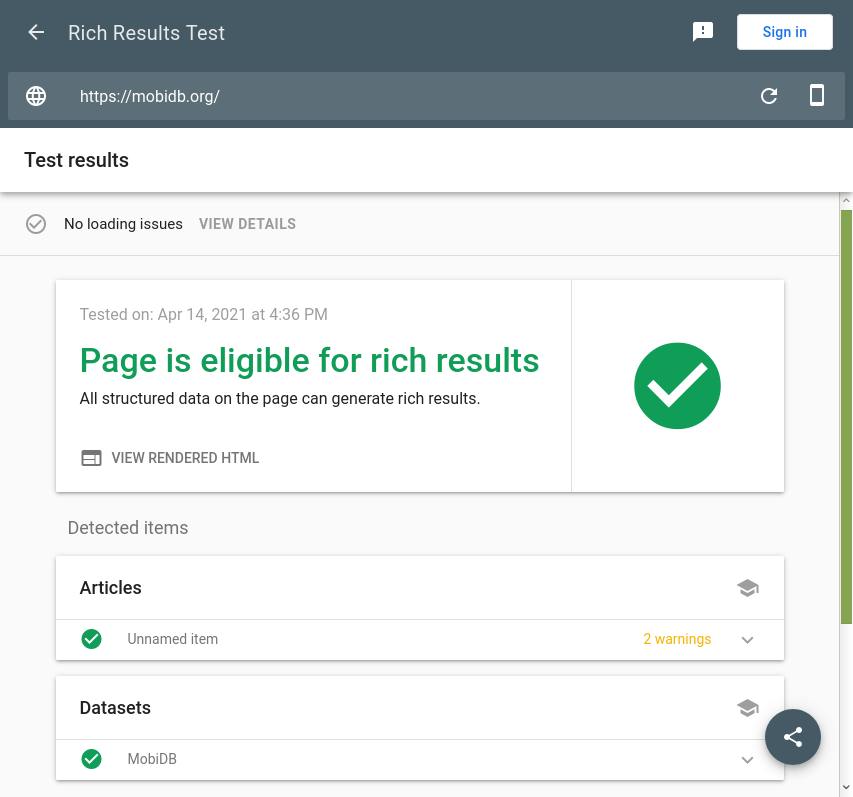
This Google tool is promoted as a successor of Google Structured Data Testing Tool and can detect Schema.org markup interpreted as “rich results” as shown on Google Search. It is very convenient for checking Datasets types. Currently it ignores Bioschemas proposals for Schema.org so it may return warnings and errors in some Bioschemas profiles.
The screenshot shows the tool results for the MobiDB database. Click here to see the results.
2.3 Bioschemas Markup Scraper and Extractor (BMUSE)
https://swel.macs.hw.ac.uk/scraper/
This is a web deployment of a tool developed by the Heriot-Watt team that is a dedicated Bioschemas scraper. It is able to deal with a wider range of Single Page Application engines than the Google tool, but does not offer much in the way of a user interface. You need to parameterise your call to the getRDF method by supplying the url and the output type option; currently we support jsonld and turtle.
For your example, to retrieve the content from the DisProt page for DP00003 we would use
https://swel.macs.hw.ac.uk/scraper/getRDF?url=https://disprot.org/DP00003&output=jsonld
2.4. Bioschemas Validator (Experimental)
http://www.macs.hw.ac.uk/SWeL/BioschemasValidator/
This service builds on the BMUSE scraper and provides a validation interface against the Bioschemas profiles.
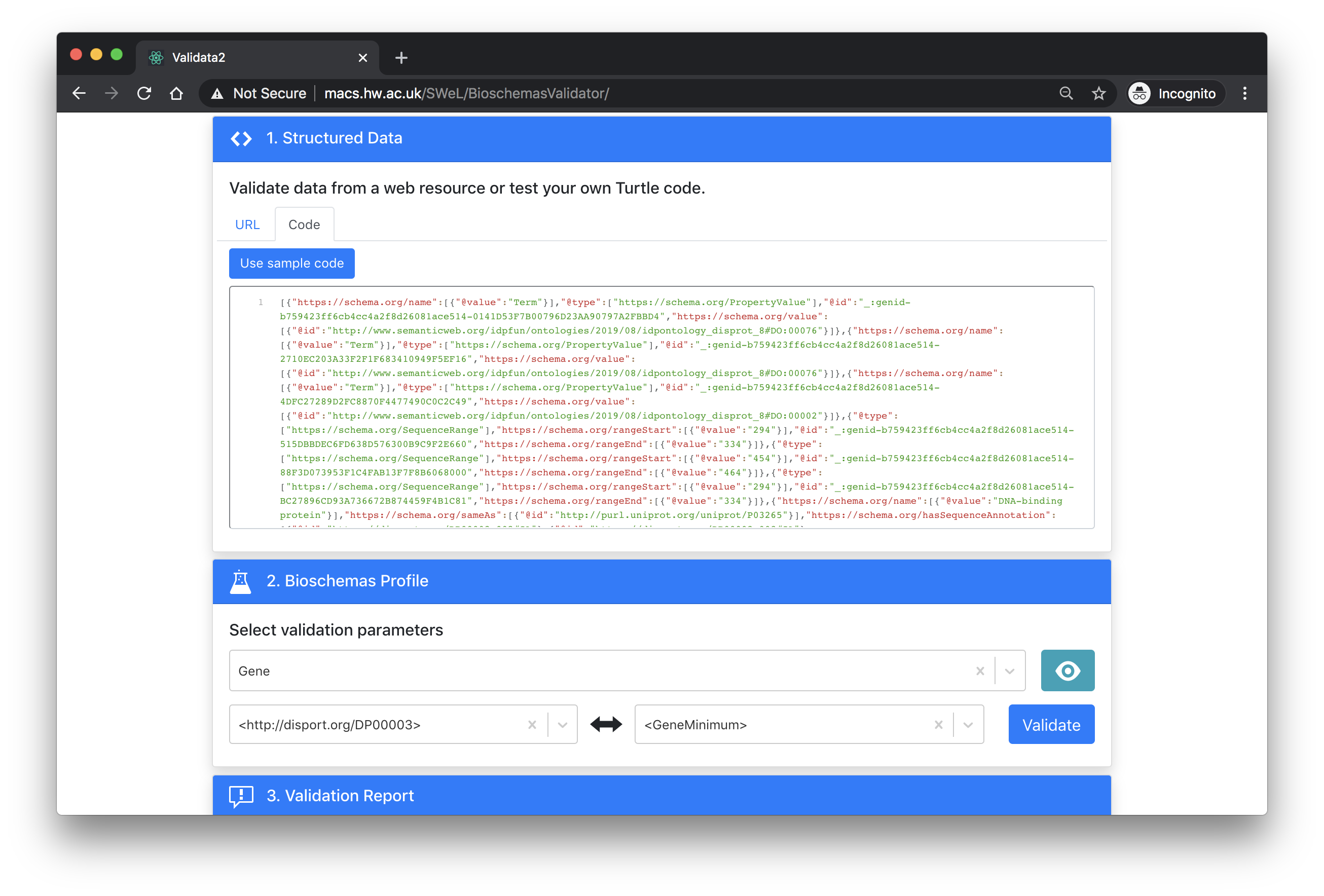
Once you have entered in your URL and loaded the content from your web page, you should select the profile that you want to validate against. The tool also needs to know which node from those detected should be validated against the profile as well as the marginality level that you want to validate at (minimal, recommended, optional).
The error report is minimal but it will give you a green or red bar to indicate success or failure.
The screenshot below shows the page for validating the DisProt protein DP00003 against the Gene profile at the minimal level. Click here to try it yourself.
Keywords: schemaorg, JSON-LD, bioschemas
Topics:
Audience:
- (Markup provider, Markup consumer) WebMaster
Authors:
Contributors:
License: CC-BY 4.0
Version: 1.1
Last Modified: 22 July 2021

