How to mark up your own resource with Bioschemas
In this how-to, we will guide you through the necessary steps in order to get a JSON-LD markup describing your own resource using a Bioschemas profile

1. Define your strategy
If your resource involves more than one profile, you first need to define the strategy that best suits your case. You can link a resource of one type, let’s say DataCatalog, to other related ones, let’s say Dataset, via properties, e.g. dataset for this case. Some properties have inverses, e.g. includedInDataCatalog, so you can go from DataCatalog to Dataset. You can choose which direction suits you best.

Link from DataCatalog to Dataset if you only have a few datasets and you introduce them all on your catalog landing page. Example: FAIRsharing (SMV)
Link from Dataset to DataCatalog if you have that many datasets; rather than list them all, you provide a search and retrieval mechanism to find them. Example: EGA (SMV), EGA Dataset WTCCC1 1958BC control dataset (SMV)
2. Map elements to properties
Now, focus on a particular profile and have at hand the profile page as well as your resource, i.e., your web pages. Do a manual exercise mapping elements in your web page to properties in the profile. This will help you have a clearer idea on what you want to achieve once you implement your markup into your website.
You might need more than one iteration here as you can face multiple options at the beginning; choose the one that gives more benefits. Keep in mind your goal for adding this mark up to your resource.
3. Add markup to your page
Now that you have identified those elements on your web page that you want to mark up, it is time to add the markup to your page. You will need to decide what format to use, how to add the markup and where to add it.
3.1. Formats
As discussed on the introduction to Schema.org, there are multiple formats that can be used to add structured markup to web pages. Bioschemas (and Schema.org) recommends JSON-LD so we will use that in the following examples.
3.2. Include standard properties
There are four properties that should always be included, these are:
@context–so it is clear what schema you are using@type–so it is clear what the type of thing you are describing@id–so it is clear what the identifier of the thing you are describing isdct:conformsTo–so it is clear what Bioschemas profile version you adhere to (note that you need to use the full IRI of the property, i.e.http://purl.org/dc/terms/conformsTo)
For instance, if you are describing a DataCatalog, the beginning of your markup would look something like:
{
"@context": "https://schema.org/",
"@type": "DataCatalog",
"@id" : "https://www.guidetopharmacology.org",
"http://purl.org/dc/terms/conformsTo": {
"@type": "CreativeWork",
"@id": "https://bioschemas.org/profiles/DataCatalog/0.3-RELEASE-2019_07_01/"
},
...
}
3.3. Markup the properties of your page
Here we have two possibilities: manually or automatically adding the markup.
Depending on how the data is rendered on your website, there might be a production pipeline behind your web pages. If this is the case, web developers will decide the best way to programmatically add the Bioschemas markup. This is commonly the case for registries/repositories or data/knowledge bases. For instance, the Ensembl Genome Browser provides information on genomes and has integrated the Bioschemas markup to their production pipeline. Thus, whenever you visit an Ensembl gene page you get not only the HTML but also the JSON-LD embedded within it, e.g. ensembl:ENSG00000139618 (SMV).
For websites without a production pipeline behind, those where you manually edit the HTML yourself, you will have to add the markup by hand. For instance, if you organize a half-day workshop with less than 10 accepted papers, you could add the corresponding markup by hand. This is the case for the Research Objects Management for Linked Open Science Workshop which includes bibliographic markup for the submissions on 2020 (SMV). The workshop web page is based on GitHub pages and the markup for the publications was manually added at the end of the corresponding MarkDown document.
3.4. Where to add the markup
You can put the JSON-LD corresponding to your markup anywhere within the HTML where a <script> element is allowed. You can choose between having multiple <script type="application/ld+json"> elements or only one. If you really need to you can even mix JSON-LD and RDFa, although we don’t recommend this as many tools will not be able to extract all your markup.
If you choose to have multiple <script type="application/ld+json"> elements, most likely you want them close to the corresponding HTML. For instance, if you visit the gene page BRCA2 at Ensembl, you will observe the following gene summary.
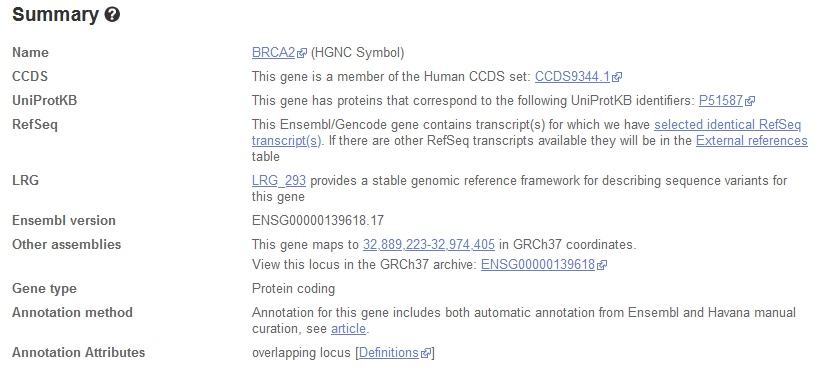
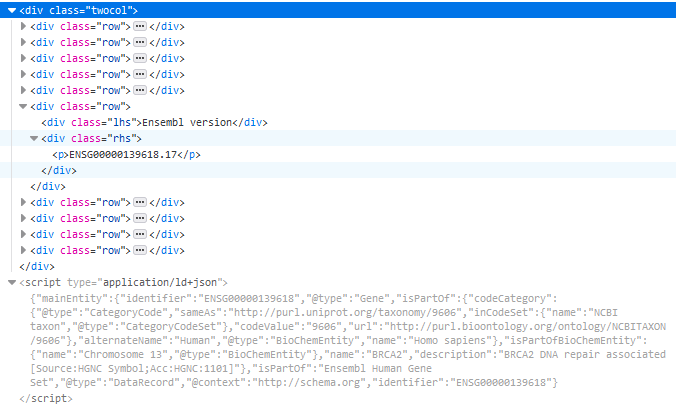
This summary corresponds to a two-colum <div> used to show the name, CCDS, UniProtKB and so on. In Figure 2, we show the respective HTML code where the two-column <div> is highlighted in blue and the row corresponding to the Ensembl version has been expanded. Below this <div> appears the JSON-LD with the corresponding markup.
If you want to include only one <script type="application/ld+json"> with all the markup relevant to your page, a common option is adding the JSON-LD at the end of the HTML code so it will not interfere with the rendering of the page. Just make sure that it is before the closing </html> tag!
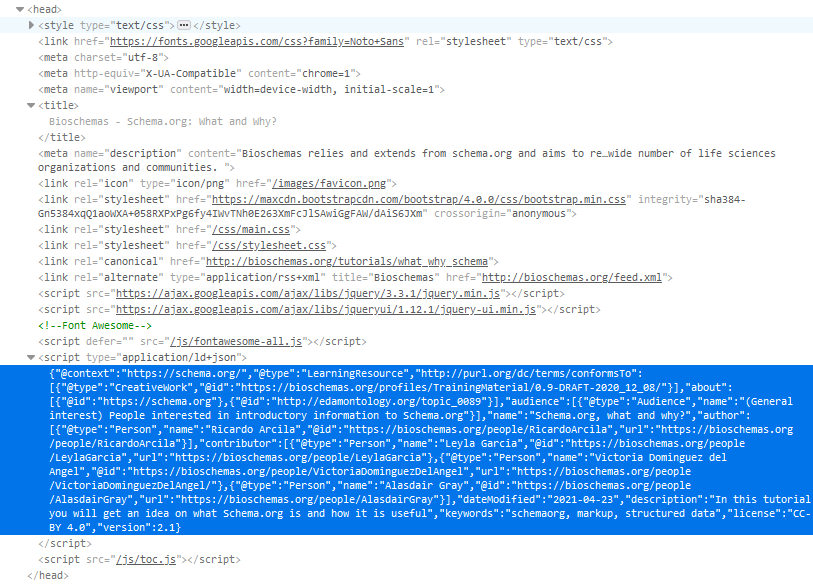
If your page is not too big, you can also choose to have the JSON-LD at the beginning of the HTML code as part of the <head>. This is the way used for Bioschemas how-to pages as shown in Figure 3 where the JSON-LD is highlighted in blue.
4. JSON-LD special characters
JSON-LD, as any other computational format, has some special characters. For example, property keys as well as property string values can be enclosed with either single or double quotations; quotation marks are therefore seen as special characters (i.e., they have a special meaning as part of the format specification and will be parsed by tools accordingly). Here you can see a key (name) - value (Bioschemas) pair, notice how the key and the value are surrounded by double quotes: "name": "Bioschemas".
If you want to include any of those special characters in either a key or a value, you will need to escape them by adding a \ before the character; for instance: "name": "\"Bioschemas\", schemas for Life Sciences" will result in the key name having the value “Bioschemas”, schemas for Life Sciences (we use here italics to make it easier to read). A special case is the \ itself; if you need to use it, you should escape it as \\. For example, this is the case with the smiles property.
Keywords: schemaorg, markup, structured data, bioschemas
Topics:
Audience:
- (Markup provider, Markup consumer) WebMaster, people interested in adding Bioschemas markup to their website
Authors:
Contributors:
License: CC-BY 4.0
Version: 3.0
Last Modified: 04 November 2021

